Background
Data on protein-drug interactions are rapidly increasing and being collected in databases such as DrugBank. The information in these databases usually reflects known/observed interactions, while the lack of data for a given protein-drug pair does not necessarily mean that those protein-drug molecules are not interacting. Indeed, recent studies supported by both computations and experiments indicate that many drugs have side effects (i.e. they target proteins) other than those known/compiled in DrugBank. This latter property may be exploited for designing ‘repurposable’ drugs or polypharmacological treatments. Efficient identification of such potential interactions is an important challenge that is likely to accelerate drug discovery and development efforts. There is a need for efficient identification of such data, and efficient dissemination of results.
Overview
In this tutorial, we will describe the following:
- The purpose of BalestraWeb
- The methodology used to build BalestraWeb
- The protocol for usage
- Programmatic access
- Drug-Target interaction prediction confidence scores and how to interpret them
- Drug-Drug and Target-Target similarity scores and how to interpret them
- The interactive network visualization
- An example use of BalestraWeb for drug repurposing
Purpose
Predicting targets of drugs is an important challenge in drug development:
- Necessary for explaining mechanism of action and side effects.
- The lack of such knowledge can cause late-phase toxicity failure.
- Enables potential fast therapy development via drug repurposing.
Method
Balestra solves this problem by learning a latent variable model of drug-target interactions by using probabilistic matrix factorization on interaction data from the DrugBank database v4 - which is the latest stable version of DrugBank at the time of writing. The latent variable model in this web-based service allows the prediction of interactions of any approved drug and any target in DrugBank. Thus, BalestraWeb transforms the DrugBank approved drug-target interaction matrix from an occupancy of 0.25% to 100%. This is due to the fact that only 4874 interactions are known between the 1,313 approved drugs and their 1,457 targets in DrugBank (hence 0.25% occupancy) meaning that users cannot find any information about 99.75% of the possible drug-target interactions - which amounts to ~1.9 million interactions. Our service predicts the likelihood of each of those interactions, thereby completing the unknown 99.75% of the interaction matrix, and makes it accessible to users.
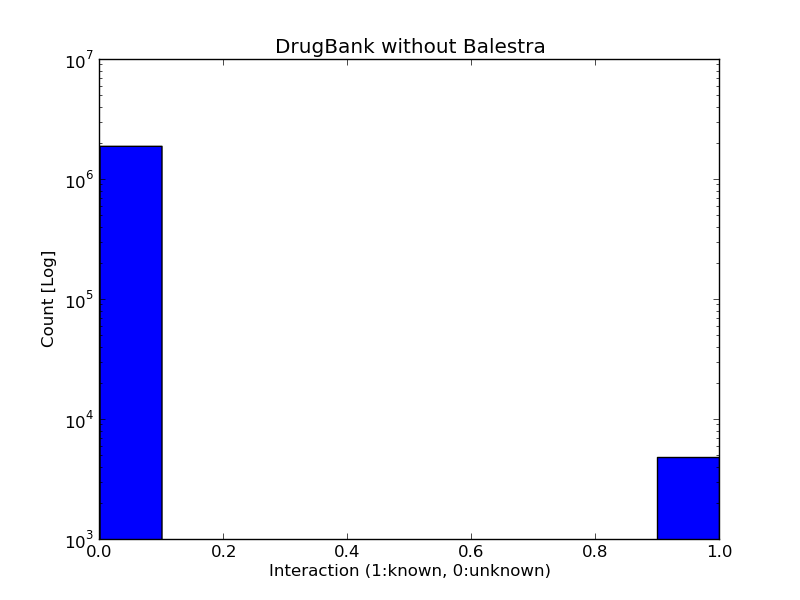
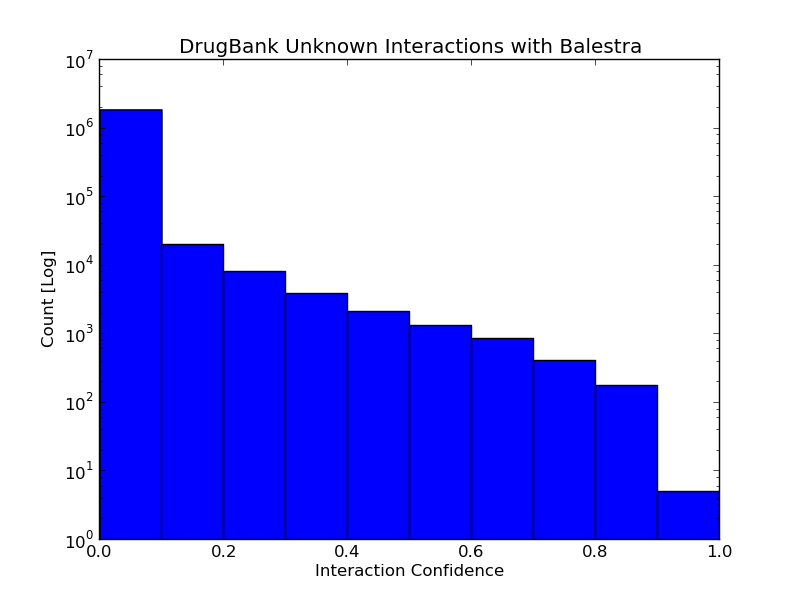
Protocol
To use the tool, please input the DrugBank identifier of any drug annotated as approved in DrugBank. These identifiers are of the form "DB12345" where the first two characters are always "DB" and the remaining numbers identify the specific drug. Identifying the drug in this manner is guaranteed to work as the DrugBank ID uniquely selects the drug of interest. To facilitate use of the service, some drug names such as "sunitinib" "atorvastatin", or "aspirin" are also recognized, however the DrugBank ID is the preferred approach.
To identify the target, the user must enter the DrugBank ID of the target of an approved drug in DrugBank. - which is the latest stable version of DrugBank at the time of building of this website. This ID is a series of numbers uniquely identifying each target in DrugBank, such as "BE0000694" for ADRB2. Alternatively the user can also enter gene identifiers from GenAtlas, or UniProt. However, since the service is built on DrugBank, the DrugBank ID of the targets of approved drugs is the preferred approach.
Having identified the drug and/or target of interest; the user has three query choices:
- Drug-Target interaction prediction,
- Drug-Drug similarity calculation,
- Target-Target similarity calculation.
For all of these queries, the user has the option to select "Secondary Interactions" when performing the query. If this option box is checked, then BalestraWeb displays the secondary interactions of all the nodes in the interactive graph as well. This enables users to quickly see possible shared drugs/targets among the known and predicted interaction partners, for example.
Drug-Target Interaction Prediction
The user can perform one of the three following operations:
- Predict interaction between a drug-target pair: The user simply inputs the drug identifier and the target identifier; and clicks the "Predict" button. The system reports the confidence with which an interaction exists between the query drug and target (more on how to interpret these confidences below) as well as all the known interactions of the drug and the target. The known interactions are displayed to allow the user to put the prediction in perspective. The reported prediction and the reported set of known interactions are also visualized as an interactive network where the user can graphically view the presented results, as well as manipulate the view. The number of predictions input box has no effect in this mode because there is only one prediction that can be made: the interaction between the query drug and target.
- Predict the most likely interaction partners of a drug: The user simply enters a drug identifier and clicks the "Predict" button. The system uses the underlying latent factor model to make predictions between the query drug and all targets. The 10 most likely interaction partners of the drug are shown by default, along with any known interactions. The user can use the "No. of Predictions" input box to change this setting to any number up to 100. An interactive network visualization of the results accompanies the result view.
- Predict the most likely interaction partners of a target: The user enters the identifier of the target of interest and clicks the "Predict" button. The mode of operation is exactly the same as described in the previous section, and the drugs most likely to interact with the query target are shown along with any known interaction partners.
Drug-Drug Similarity Calculation
The user can perform the two following operations:
- Calculate similarity between two drugs: The user enters the identifier of the two drugs of interest and clicks the "Predict" button. The similarity between the two input drugs based on their latent variable feature vectors is shown along with all known interactions of the two drugs. The interactions of the drugs as well as the similarity between them is shown in an interactive network visualization. Again, since the user is asking for exactly one prediction, the no. of predictions input box is irrelevant.
- Find the drugs most similar to a query drug: The user enters the identifier of the query drug and clicks the "Predict" button. The similarity between the latent variable descriptor of the query drug and all other drugs is calculated; the drugs most similar to the query drug are returned. By default the top ten most similar drugs are returned however the user can choose a different value by entering it in the "no. of predictions" box.
Target-Target Similarity Calculation
The operation mechanism is identical to the drug-drug similarity calculation discussed above, using target identifiers instead of drug identifiers.
Programmatic Access
BalestraWeb uses HTML primitives by design for cross-platform stability and reliability. One of the advantages of this design decision is that automated browsing (for hundreds or thousands of requests) is very easy. Using one of many automated web browsing packages, such as mechanize and a scripting language like Python the user can programmatically access BalestraWeb only in a few lines of code. We provide a demo script that does this (available here) which the users can modify according to their needs. Alternatively, the users can download the entire code and auxiliary files that run BalestraWeb here.
Interpreting Confidence Scores for Drug-Target Interactions
The reported interaction confidence, as expected, has a large range. To put these confidence scores into context the following guide can be used:
- >90%: Very High Confidence
- 80% to 90%: High Confidence
- 60% to 80%: Medium Confidence
- <60%: Low Confidence
Interpreting Drug-Drug and Target-Target Similarity Scores
We compute the drug-drug and target-target similarities using
cosine similarity; therefore
the values range between -1 (anti-correlated) to 0 (no similarity) to 1 (highly similar).
The similarities between all drug-drug and target-target pairs are distributed as shown
in the histograms below.
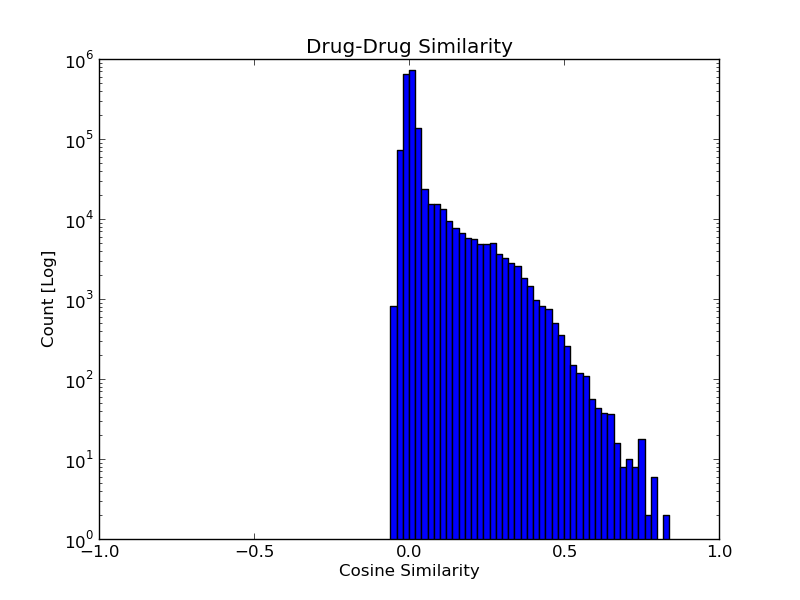
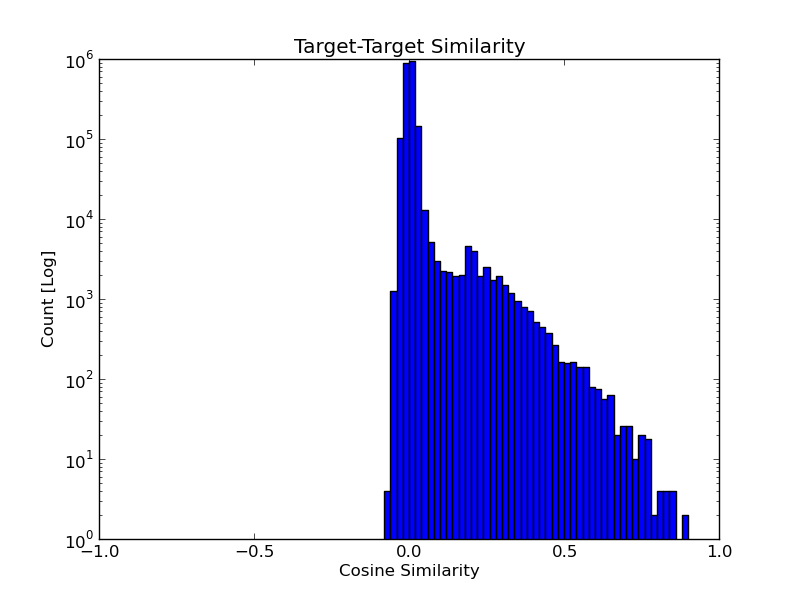 When analyzing these similarity scores, the user can refer to the following guide:
When analyzing these similarity scores, the user can refer to the following guide:
- >0.65: High Similarity
- 0.55 to 0.65: Medium Similarity
- -0.1 to 0.55: Low Similarity
- <-0.1: Anti-Correlation
Interactive Network Visualization
Whenever results are presented to the user, there is an interactive network visualization that accompanies. These network visualizations serve to make it easy for the user to visually inspect the results and form an idea about the network in one look. Therefore the node links represent the type of interaction between the linked nodes using the same color coding as the results table: red indicates predicted interactions, gray indicates known interactions. The thickness of the edge indicates the confidence of the shown interaction. The color of the nodes denote whether if the node represents a drug (red) or target (blue). If the user wishes to download the shown network for local processing, (s)he can click the "Download Network" link that appears next to each interactive network to download the network as a JSON data exchange format file. Almost every programming language has a JSON parser which can be found, along with a plethora of other information, at the JSON homepage.
Example: Drug Repurposing against GBRT
In this section, we will demonstrate the use of BalestraWeb through the use of an example. Essentially, the concepts and methods described in the previous sections will be utilized within the framework of a concrete example to enable the user to better grasp the use of BalestraWeb. We will setup the example as follows: the user aims to modulate the function of GBRT (GABA receptor subunit theta). This might be motivated by the fact that the protein has been implicated to be integral in a pathological process; or to interrogate the system of interest when GBRT modulation can influence the pathway of interest, to give a few examples.
Without BalestraWeb, the user would be forced to look at various databases and find only a few known drug interactions for GBRT: DrugBank lists eight, four approved and four illicit drugs (as shown here). With BalestraWeb, the user simply goes to the BalestraWeb main page, enters "GBRT" to the target box and clicks the "Predict" button. The user can immediately discover the top 10 drugs predicted to be the most likely interaction partners of GBRT - two drugs (cinolazepam and adinazolam) predicted with around 70% confidence.
Based on this information, the user can choose to look for other drugs similar to cinolazepam. BalestraWeb can be used for this purpose as well: user simply clicks the Drug-Drug button on the navigation bar, and enters "cinolazepam". On this page, there are two input boxes: one for the main drug of interest, the second box for the optional second drug to compute similarity to the primary drug. For drug-drug similarity queries, BalestraWeb calculates the cosine similarity of the latent variable vectors of the drugs. If there is only one input, BalestraWeb reports the top n (10 by default, user adjustable) most similar drugs. If the user inputs two drugs, BalestraWeb computes the similarity between these two drugs and reports it along with the targets of these drugs. Since the similarity is calculated using a latent variable model trained from drug-target interactions, this way of computing drug-drug similarity is fundamentally different than the 2D or 3D structure based comparison. Returning to our example, cinolazepam (which is an anti-depressant) has been found to have the highest similarity to adinazolam (also an anti-depressant). The second most similar drug is oxazepam (which is an anti-anxiety agent). As highlighted by these examples, the latent variable can find therapeutically similar drugs and is therefore capable of providing researchers with new directions to pursue in their research. The exact same functionality exists for targets as well and can be used by clicking the Target-Target button.